Weakly Supervised Learning with Positive and Unlabeled Data for Automatic Brain Tumor Segmentation
Applied Sciences 2022
Abstract
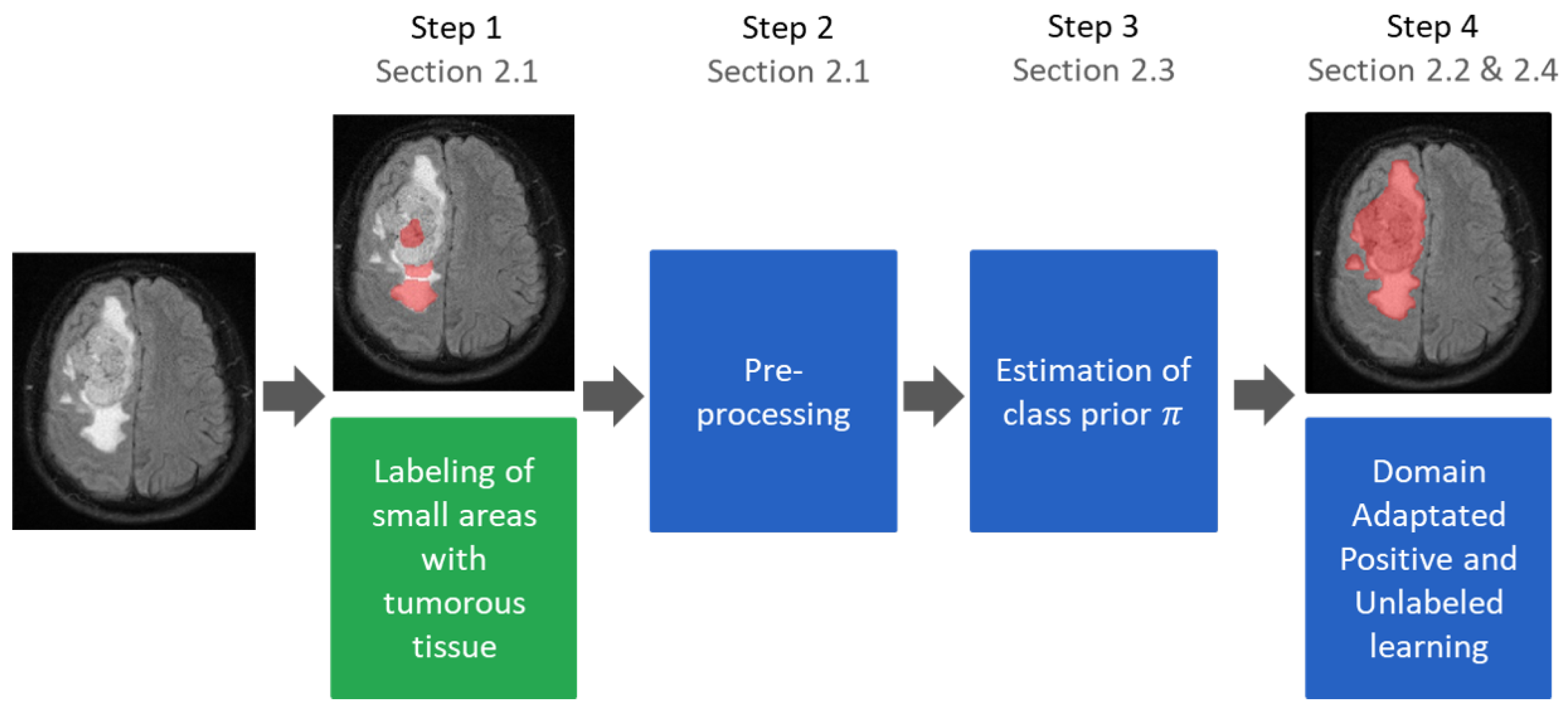
A major obstacle to the learning-based segmentation of healthy and tumorous brain tissue is the requirement of having to create a fully labeled training dataset. Obtaining these data requires tedious and error-prone manual labeling with respect to both tumor and non-tumor areas. To mitigate this problem, we propose a new method to obtain high-quality classifiers from a dataset with only small parts of labeled tumor areas. This is achieved by using positive and unlabeled learning in conjunction with a domain adaptation technique. The proposed approach leverages the tumor volume, and we show that it can be either derived with simple measures or completely automatic with a proposed estimation method. While learning from sparse samples allows reducing the necessary annotation time from 4 h to 5 min, we show that the proposed approach further reduces the necessary annotation by roughly 50% while maintaining comparative accuracies compared to traditionally trained classifiers with this approach.
Bibtex
@article{wolf2020weakly,
title={Weakly Supervised Learning with Positive and Unlabeled Data for Automatic Brain Tumor Segmentation},
author={Wolf, Daniel and Regnery, Sebastian and Tarnawski, Rafal and Bobek-Billewicz, Barbara and G{\"o}tz, Michael},
year={2022},
journal={Applied Sciences},
doi={https://doi.org/10.3390/app122110763}
}