Automatic identification of crossovers in cryo-EM images of murine amyloid protein A fibrils with machine learning
Journal of Microscopy 2020
Abstract
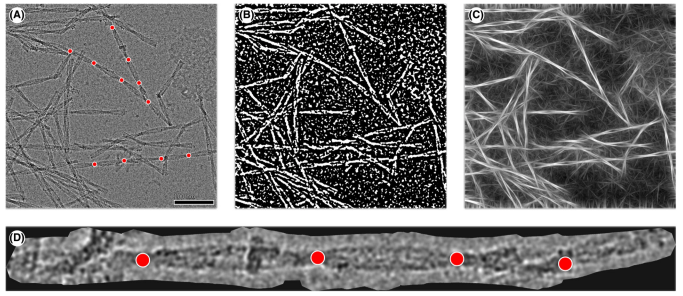
Detecting crossovers in cryo-electron microscopy images of protein fibrils is an important step towards determining the morphological composition of a sample. Currently, the crossover locations are picked by hand, which introduces er- rors and is a time-consuming procedure. With the rise of deep learning in computer vision tasks, the automation of such problems has become more and more applicable. However, because of insufficient quality of raw data and missing labels, neural networks alone cannot be applied successfully to target the given problem. Thus, we propose an approach combining conventional computer vision techniques and deep learning to automatically detect fibril crossovers in two-dimensional cryo-electron microscopy image data and apply it to murine amyloid protein A fibrils, where we first use direct image pro- cessing methods to simplify the image data such that a con- volutional neural network can be applied to the remaining segmentation problem.
Bibtex
@article{weber2020automatic,
title={Automatic identification of crossovers in cryo-EM images of murine amyloid protein A fibrils with machine learning},
author={Weber, Matthias and B{\"a}uerle, Alex and Schmidt, Matthias and Neumann, Matthias and F{\"a}hndrich, Marcus and Ropinski, Timo and Schmidt, Volker},
year={2020},
journal={Journal of Microscopy},
volume={277},
pages={12--22},
issue={1},
doi={10.1111/jmi.12858}
}