Intrinsic-Extrinsic Convolution and Pooling for Learning on 3D Protein Structures
International Conference on Learning Representations 2021
Abstract
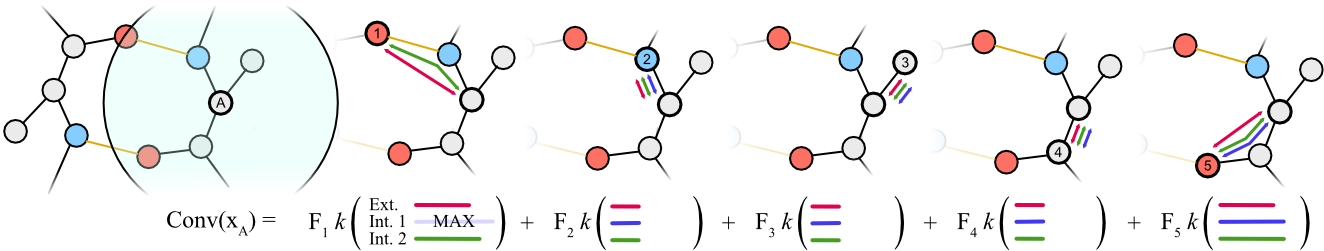
Proteins perform a large variety of functions in living organisms and thus play a key role in biology. However, commonly used algorithms in protein learning were not specifically designed for protein data, and are therefore not able to capture all relevant structural levels of a protein during learning. To fill this gap, we propose two new learning operators, specifically designed to process protein structures. First, we introduce a novel convolution operator that considers the primary, secondary, and tertiary structure of a protein by using n-D convolutions defined on both the Euclidean distance, as well as multiple geodesic distances between the atoms in a multi-graph. Second, we introduce a set of hierarchical pooling operators that enable multi-scale protein analysis. We further evaluate the accuracy of our algorithms on common downstream tasks, where we outperform state-of-the-art protein learning algorithms.
BibTeX
@inproceedings{hermosilla2021proteins,
title={Intrinsic-Extrinsic Convolution and Pooling for Learning on 3D Protein Structures},
author={Hermosilla, Pedro and Sch{\"a}fer, Marco and Lang, Matej and Fackelmann, Gloria and V{\'a}zquez, Pere-Pau and Kozl{\'i}kov{\'a}, Barbora and Krone, Michael and Ritschel, Tobias and Ropinski, Timo},
booktitle={Proceedings of International Conference on Learning Representations}
year={2021}
}